Expression Datasets
01 Picual all tissues
RNA-seq of roots, stem, meristem, leaves, flowers, pollen, fruits (epicarp + mesocarp), seeds, embryo, and testa+endosperm, combined from several experiments.
Experimental Conditions (all in control conditions with 2 replicates unless otherwise noted):
- Roots.
- Roots 4 months.
- Stem.
- Meristem.
- Leaves.
- Leaves 4 months.
- Flowers: Recently bloomed whole flowers (at anthesis).
- Pollen mature: 3 replicates of mature pollen.
- Pollen germ 6h: 3 replicates of pollen after 6 hours of germination.
- Fruits: Reaching the turning ripening state, veraison.
- Seed: Mature seed (whole), 1 replicate.







From left to right are shown figures of Picual 4 month old roots, stem, meristem, leaves, 4 month old leaves, flowers, rehydrated mature pollen, fruits, seeds, seed testa + endosperm, and embryos.
Leaves and Roots (in green) are control samples from 4 month old potted plants in conditions as published by Leyva-Pérez et al., 2015. Seed (in blue) is a control sample. Pollen samples correspond to mature rehydrated pollen, and Pollen germ 6h correspond to pollen after 6 hours of germination as published by Bullones et al., 2023. The rest of the tissues (in black) come from three healthy 10-year-old olive trees under field conditions. They were published by Ramírez-Tejero et al., 2020.
All samples were normalized to TPM.
02 Picual plant organs
RNA-seq of roots, stem, meristem, leaves, flowers, and fruits (epicarp and mesocarp) of three healthy 10-year-old olive trees under field conditions. All the samples were collected at the same time, except for fruits, which had to be collected later upon reaching the turning ripening state.
Experimental Conditions (all in control conditions with 2 biological replicates):
- Roots
- Stem
- Meristem
- Leaves
- Flowers: Recently bloomed whole flowers (at anthesis).
- Fruits: Reaching the turning ripening state, veraison.





From left to right are shown figures of Picual stem, meristem, leaves, flowers, and fruits.
The plant samples employed for this study were obtained from the World Olive Germplasm Collection (WOGC) of the Andalusian Institute of Agricultural and Fisheries Research and Training (IFAPA). Two biological replicates per sample were sequenced, and each biological replicate consisted in an equilibrated pool of three plant RNAs. cDNA libraries were sequenced by PE sequencing (100 x 2) with an Illumina HiSeq 2000 sequencer and normalized to TPM.
This dataset was published by Ramírez-Tejero et al., 2020 and raw data can be found in the BioProject PRJNA590386.
03 Picual pollen germination
RNA-seq of pollen tube growth at four times of development. Pollen grains were pre-hydratated by incubation in a humid chamber at room temperature for 1 hour and then transferred to Petri dishes containing germination medium. Petri dishes were maintained at room temperature in the dark and under gentle agitation. Pollen grains were sampled at 1, 3 and 6 hours after transfer to the in vitro germination medium.
Experimental Conditions (3 biological replicates):
- Pollen mature. Mature rehydrated pollen.
- Pollen germ 1h. Pollen after 1 hour of germination.
- Pollen germ 3h. Pollen after 3 hours of germination.
- Pollen germ 6h. Pollen after 6 hours of germination.

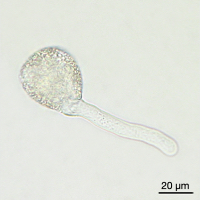

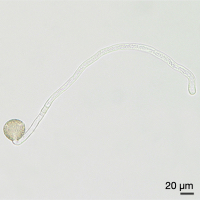
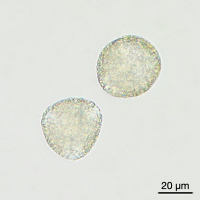
Images of Picual pollen grains at different stages of in vitro germination. From left to right, mature rehydrated pollen, and germinated pollen at 1, 3 and 6 hours of culture.
Mature pollen grains were collected during the anthesis of adult olive trees growing at Estación Experimental del Zaidín in Granada, Spain. Three biological replicates per sample were sequenced and normalized to TPM.
This dataset was published by Bullones et al., 2023.
04 Picual whole seed
RNA-seq of mature seeds sampled from ripe olives. Fruits were manually destoned and seeds were extracted by mechanical breakage of the woody endocarp. The seed tissues (testa + endosperm and embryo) were dissected with a scalpel.
Experimental Conditions (3 biological replicates):
- Seeds. Mature seed (whole), 1 replicate.
- Testa+Endosperm. Endosperm and seed coat (testa), 3 replicates.
- Embryo. Mature embryo, 3 replicates.

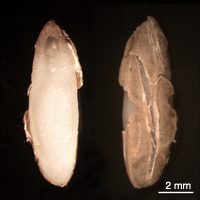

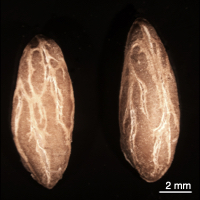
Images of Picual seeds. Form left to right, whole mature seeds, seed coat (testa) and endosperm tissues, and mature embryos.
Olive fruits were collected 210 days after anthesis from adult olive trees growing at the Estación Experimental del Zaidín in Granada, Spain. One biological replicate of Seed and three biological replicates of Testa+Endosperm and Embryo were sequenced. Data shown were normalized to TPM.
Note: This dataset is unpublished and the manuscript is in preparation. The gene expression data of endosperm and embryo samples will be released after publication.
05 Picual stresses
RNA-seq of leaves and roots from 4 month old potted olive plants in control and stess conditions. Abiotic stresses consisted of wounded roots and low temperatures, and biotic estress of infection with the fungal plant pathogen Verticillium dahliae.
Verticillium infection was performed by root-dip inoculation in a conidia suspension of a Verticillium dahliae isolate representative of the so-called defoliating pathotype. Cold stress conditions were 10°C during the day and 4°C in the night, with 14 hours photoperiod.
Experimental Conditions (2 biological replicates):
- Leaves control.
- Leaves Verticillium 15d. Leaves infected with Verticillium for 15 days.
- Leaves root wound 15d. Leaves of plants with wounds in roots, after 15 days.
- Leaves cold 24h. Leaves after 24 hours in cold conditions.
- Leaves cold 10d. Leaves after 10 days in cold conditions.
- Roots control.
- Roots Verticillium 48h. Roots infected with Verticillium for 48 hours.
- Roots Verticillium 7d. Roots infected with Verticillium for 7 days.
- Roots Verticillium 15d. Roots infected with Verticillium for 15 days.
- Roots wound 8h. Roots wounded, after 8 hours.
- Roots wound 24h. Roots wounded, after 24 hours.
- Roots wound 48h. Roots wounded, after 48 hours.
- Roots wound 7d. Roots wounded, after 7 days.




4 month old Picual plants in stress conditions, from left to right: Leaves control, leaves in cold stress conditions for 24 hours, leaves in cold stress conditions for 10 days infected with Verticillium, and roots in control condition.
The plant samples employed for this study were obtained from a commercial nursery located in Córdoba province, Spain. Two biological replicates per sample were sequenced. cDNA libraries were sequenced by PE sequencing (100 x 2) with an Illumina HiSeq 1000 sequencer and normalized to TPM.
This dataset was published by Leyva-Pérez et al. 2014 and raw data can be found in the BioProject PRJNA256033.
06 Picual cold stress
RNA-seq of leaves from 4 month old potted olive plants in control and low temperature stess conditions, with 10°C during the day and 4°C in the night, with 14 hours photoperiod.
Experimental Conditions (2 biological replicates):
- Leaves control. (A).
- Leaves cold 24h. Leaves after 24 hours in cold conditions. (B).
- Leaves cold 10d. Leaves after 10 days in cold conditions. (F).

(A) Control plant; (B) cold-stressed plant during 1 day; (C) cold-stressed plant during 2 days; (D) cold-stressed plant during 3 days; (E) almost fully recovered plant after 6 days of cold stress; (F) fully recovered plant after 10 days of cold stress. Plants show flaccid leaved and stresed phenotype in B and is progessively recovering from D to F.
The plant samples employed for this study were obtained from a commercial nursery located in Córdoba province, Spain. Two biological replicates per sample were sequenced. cDNA libraries were sequenced by PE sequencing (100 x 2) with an Illumina HiSeq 1000 sequencer and normalized to TPM.
This dataset was published by Leyva-Pérez et al. 2014 and raw data can be found in the BioProject PRJNA256033.
07 Picual wound stress
RNA-seq of leaves and roots from 4 month old potted olive plants in control conditions and with wounded roots.
Experimental Conditions (2 biological replicates):
- Leaves control.
- Leaves root wound 15d. Leaves of plants with wounds in roots, after 15 days.
- Roots control.
- Roots wound 8h. Roots wounded, after 8 hours.
- Roots wound 24h. Roots wounded, after 24 hours.
- Roots wound 7d. Roots wounded, after 7 days.


4 month old Picual plants in control conditions, leaves on the left and roots on the right.
The plant samples employed for this study were obtained from a commercial nursery located in Córdoba province, Spain. Two biological replicates per sample were sequenced. cDNA libraries were sequenced by PE sequencing (100 x 2) with an Illumina HiSeq 1000 sequencer and normalized to TPM.
This dataset was published by Leyva-Pérez et al. 2014 and raw data can be found in the BioProject PRJNA256033.
08 Picual Verticillium infection
RNA-seq of leaves and roots from 4 month old potted olive plants in control conditions and infected with the fungal plant pathogen Verticillium dahliae. The infection was performed by root-dip inoculation in a conidia suspension of a Verticillium dahliae isolate representative of the so-called defoliating pathotype.
Experimental Conditions (2 biological replicates):
- Leaves control.
- Leaves Verticillium 15d. Leaves infected with Verticillium for 15 days.
- Roots control.
- Roots Verticillium 48h. Roots infected with Verticillium for 48 hours.
- Roots Verticillium 7d. Roots infected with Verticillium for 7 days.
- Roots Verticillium 15d. Roots infected with Verticillium for 15 days.


Picual plants in control conditions (left) and infected with Verticillium (right).
The plant samples employed for this study were obtained from a commercial nursery located in Córdoba province, Spain. Two biological replicates per sample were sequenced. cDNA libraries were sequenced by PE sequencing (100 x 2) with an Illumina HiSeq 1000 sequencer and normalized to TPM.
This dataset was published by Leyva-Pérez et al. 2014 and raw data can be found in the BioProject PRJNA256033.
09 Roots of olive cultivars with variable tolerance to Verticillium
RNA-seq of roots from 14 to 20 years old olive plants corresponding to 36 olive cultivar in good phytosanitary conditions without any disease symptoms.
Cultivars grouped by their resistance or susceptibility to the fungal plant pathogen Verticillium dahliae (2 biological replicates):
- Highly Resistant: Frantoio, Manzanillera de Huercal Overa.
- Resistant: Dokkar, Koroneiki, Leccino, Maarri, Maureya, Uslu.
- Moderately Susceptible: Arbequina, Barnea, Barri, Fishomi, Klon 14, Manzanilla de Sevilla. Lianolia Kerkyras, Morrut, Myrtolia, Picual, Picudo, Piñonera, Verdial de Vélez Málaga.
- Susceptible: Abbadi Abou Gabra, Abou Salt Mohazam, Chemlal de Kabylie, Llumeta, Menya, Temprano.
- Extremely Susceptible: Jabali, Mastoidis.
- Not clear phenotype: Abou Kanani, Forastera de Tortosa, Grappolo, Kalamon, Majhol-1013, Majhol-152, Mari, Megaritiki y Shengeh.
Root samples of 58 adult plants corresponding to 29 olive cultivars (two biological replicates each) were collected at the World Olive Germplasm Bank at IFAPA Centro ‘Alameda del Obispo’ located in Córdoba province, Spain. The olive trees under study were all adults, in productive stage and in good phytosanitary conditions without any disease symptoms. Their age ranged from 14 to 20 years old. They were randomly planted at 7 × 7 m spacing in alkaline loam and sandy-loam soil. Samples were obtained around 30 to 40 cm from the trunk. Two biological replicates per sample were sequenced. cDNA libraries were sequenced by PE sequencing (100 x 2) with an Illumina HiSeq 2500 sequencer and normalized to RPKM.
This dataset was published by Ramírez-Tejero et al. 2021 and raw data can be found in the BioProject PRJNA638671.
10 Souri drought
RNA-seq of wood from branches of twelve 3-year-old olive plants corresponding to Souri cultivar in pots under 90 days of seasonal spring to summer warming. Six trees out of twelve were exposed to limited water conditions for 28 days. The six watered trees were irrigated continuously. Samples were collected at four time-points: day -3 (baseline), day 13 (heat wave and early drought), day 27 (late drought) and day 80 (post-drought).
Experimental Conditions (3 biological replicates):
- Watered trees -3d.
- Watered trees 13d.
- Watered trees 27d.
- Watered trees 80d.
- Dried trees -3d.
- Dried trees 13d.
- Dried trees 27d.
- Dried trees 80d.
The plant samples employed in this study were collected at the Weizmann Institute of Science located in Rehovot, Israel. Three biological replicates per sample were sequenced. cDNA libraries were sequenced by PE sequencing (100 x 2) with an Illumina HiSeq 2500 sequencer and normalized to RPKM.
This dataset was published by Tsamir-Rimon et al. 2020 and raw data can be found in the BioProject PRJNA606032.


